Abstract
Monosomy 7 or the loss of the long arm of chromosome 7 (-7q) are prevalent cytogenetic abnormalities in pediatric myeloid neoplasms, occurring in myelodysplastic syndromes (MDS, ~30% of cases), therapy-related myeloid neoplasms (tMN, ~30%), juvenile myelomonocytic leukemias (JMML, ~25%), acute myeloid leukemias (AML, ~5%), among others. These chromosomal deletions are associated with a high risk of disease progression and poor outcome, presumably resulting from haploinsufficiency of key hematopoietic regulators on chromosome 7, such as SAMD9, SAMD9L, KMT2C/MLL3, EZH2, and DOCK4. Characterization of these aberrations currently relies on cytogenetics and genomic analysis. Recently, gene expression at the cellular level has been explored in the myeloid compartment of patients with monosomy 7, however, the progression of monosomy 7 at a single-cell level across all cell types in pediatric disease has yet to be investigated.
To address this, we employed a combination of whole genome sequencing (WGS), whole exome sequencing (WES), and/or custom targeted-capture sequencing on a cohort of 122 pediatric and young adult patients with chromosome 7 abnormalities (age range 0.5-24.6, median 6.5, average 8.5) to define single nucleotide variations (SNV), insertion/deletion events (InDel), structural variations (SV), and copy number alterations (CNA). This cohort represents a variety of diagnoses including AML (44%), MDS (22%), tMN (17%), JMML (2%), acute leukemia of ambiguous lineage (1%) and unclassified myeloid neoplasms (14%). We identified various disruptions to chromosome 7, including complete loss (62%), full or partial loss of 7q (25%), copy-neutral loss of heterozygosity (CN-LOH, 4%), gain of chr7/7q (5%), and complex changes including both gain and loss on the same chromosome (4%). Our data identified a set of commonly mutated genes including PTPN11 (24%), KRAS (16%), NRAS (15%), RUNX1 (14%), NF1 (13%), ASXL1 (12%), ETV6 (11%), SETBP1 (11%), EZH2 (9%) and TP53 (7%).
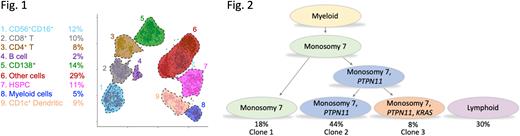
The genomic information compiled for each patient allowed us to choose 8 representative samples to undergo single-cell proteogenomic multi-omics (scDNA-Ab-seq) using Tapestri reagents (MissionBio). This platform enables the simultaneous capture of DNA genotype and cell-surface protein immunophenotype from a single cell. We designed a custom DNA panel to detect recurrent mutations in pediatric myeloid neoplasms, including the entire coding region of SAMD9 and SAMD9L which are known to correlate with monosomy 7. Simultaneously, we used a comprehensive protein panel capable of detecting 42 unique cell surface antigens to phenotypically define cell populations (Fig. 1). Merging the genotype and immunophenotype information enabled us to precisely determine the pathogenic trajectories and describe clonal evolution. In this example, loss of chromosome 7 and the presence of a recurring PTPN11 p.D61G SNV is restricted to the immature myeloid population. The monosomy 7 clone first acquired a PTPN11 mutation, then subsequently acquired a KRAS mutation, creating a third, minor clone harboring both PTPN11 and KRAS (Fig. 2).
Collectively, we comprehensively mapped chromosome 7 loss across diverse pediatric myeloid neoplasms. Using single-cell multi-omics, we determined that monosomy 7 is the initiating event for each of the studied patients. We were also able to identify co-occurring mutations, their cell of origin, and the trajectories of the clonal expansion. Our data has clarified the progression and pathogenesis of monosomy 7 and shed light on the complexity of the associated catastrophic diseases.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal